This is the fourth edition of this visualisation, previous editions were in June 2014, October 2013 and December 2012.
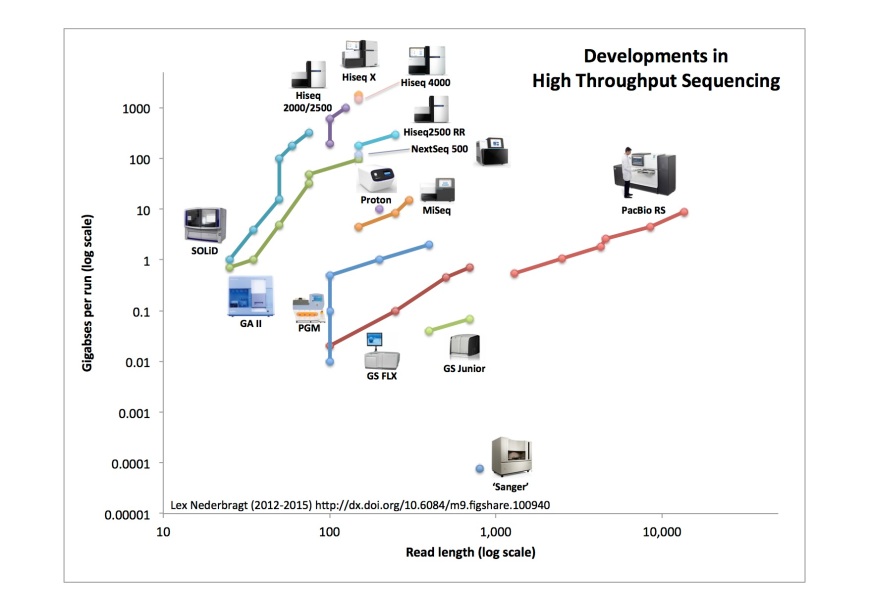
As before, full run throughput in gigabases (billion bases) is plotted against single-end read length for the different sequencing platforms, both on a log scale. Yes, I know a certain new instrument seems to be missing, hang on, I’m coming back to that…
Notable changes from the June 2014 edition
- I added the Illumina HiSeq 4000
- the HiSeq 2500 Rapid Run upgraded to 2×250 bp read length
- PacBio upgraded to P6-C4 metrics
- read numbers (but not the other metrics) for the full PacBio runs were updated, as they previously reflected those for single SMRTCells
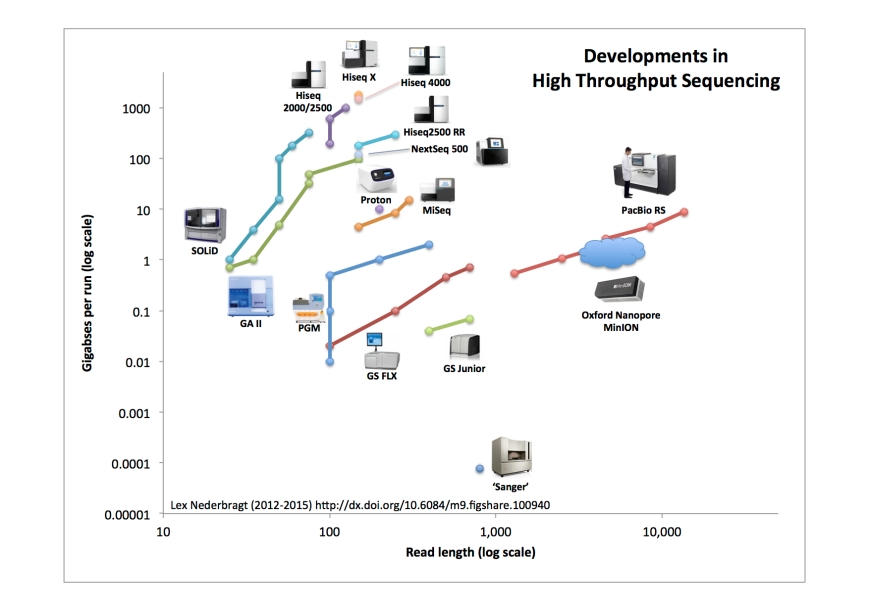
But, where is the Oxford Nanopore MinION?
The Oxford Nanopore MinION is a bit tricky. My metrics are based on company specifications that anyone can view from their website, for commercially released instruments and (chemistry) updates. The commercial release of the MinION seems now to have happened, as it was announced everyone can apply for the MinIon Access Program and will be accepted (barring some sanity screening they probably will do). But metrics for this instrument are a different matter. There are no company specs that I can find. Partly this is understandable, as read length, for example, is dependent on the input length of the sample (or library, rather). The other reason for the lack of specifications may be the Minion Access Programs philosophy of Oxford Nanopore. It is the users that are discovering what the instrument can do, rather than the company telling the customer what to expect.
For my visualisation, this lack of specification causes a problem, as this makes it impossible to point to a source for where to place the MinION on the plot. Hence it is lacking from the figure above.
As a (temporary?) solution, here is, purely based on what I read in articles, both published scientific articles, preprints, and webpages, where the MinION more or less belongs. Note that I choose to represent it with a cloud rather than a single, solid datapoint…
Some comments
- note how close together the data points fall for the HiSeq 4000 and the HiSeq X
- [EDIT] another new ‘instrument’ is the HiSeq X Five, but the single instrument is the same as for the HiSeq X Ten, and so now new datapoint was generated for the X Five
- Illumina instruments clearly dominate the top middle part of the figure (between 100 and 300 bp, and 10 GB and 2 TB throughput)
- the Complete Genomics/BGI Revolocity is missing from the figure, also since there are no company specifications on the Complete Genomics website. There is as of yet not enough information to try to piece together what the metrics are, all that is known is that the read length is 28 bp paired end.
- [EDIT] a reader left a comment which I’d like to quote: “The Amersham/GE Healthcare MegaBACE 4500 was a 384-capillary instrument, with readlengths over 1000 bp. Yet, due to ABI’s better dominance in the market, the MB 4500 never had much penetration.”
- [EDIT] see also this reader comment on the history of the ABI 37* instruments. I’ve added 2002 as release date for the 3730xl as a result. It would be fun to try to dig up more information on the other instruments on the market before the 454 and Solexa ones came out…
- as mentioned in the original blog post: some data was obtained by going to previous versions of company websites through the Internet Archive
- I used full single-run specs with maximally stated throughput as available at the time of writing
- sometimes, the total numbers of reads per full run and total bases obtained do not match up; for the figure, I always chose the reported throughput in bases
- for Illumina, I chose to use the single-end read length, although the maximum throughput was based on the sum of all reads from a paired end run; I felt it unfair to double the read length for this platform for the figure
Availability
Data and figures are released under a CC0 license at figshare, with doi 10.6084/m9.figshare.100940. I’ve also added the content to Github at https://github.com/lexnederbragt/developments-in-next-generation-sequencing.
Disclaimer
As before: although I took utmost care in collecting the data, I may have gotten some of my numbers completely wrong, for which I apologise in advance; please help me correct any mistakes or omissions through leaving a comment, or sending me a pull request.
Finally, the raw data
| Platform | Instrument | Year | Reads per run | Read length (mode or average) | Bases per run (gigabases) | Source |
|---|---|---|---|---|---|---|
| ABI Sanger | 3730xl | 2002 | 96 | 800 | 0.0000768 | 0 |
| 454 | GS20 | 2005 | 200000 | 100 | 0.02 | |
| 454 | GS FLX | 2007 | 400000 | 250 | 0.1 | |
| 454 | GS FLX Titanium | 2009 | 1000000 | 500 | 0.45 | |
| 454 | GS FLX+ | 2011 | 1000000 | 700 | 0.7 | 1 |
| 454 | GS Junior | 2010 | 100000 | 400 | 0.04 | 2 |
| 454 | GS Junior+ | 2014 | 100000 | 700 | 0.07 | 16 |
| IonTorrent | PGM 314 chip | 2011 | 100000 | 100 | 0.01 | 3 |
| IonTorrent | PGM 316 chip | 2011 | 1000000 | 100 | 0.1 | 3 |
| IonTorrent | PGM 318 chip | 2011 | 5000000 | 100 | 0.5 | 3 |
| IonTorrent | PGM 318 chip | 2012 | 5000000 | 200 | 1 | 3 |
| IonTorrent | PGM 318 chip V2 | 2013 | 5000000 | 400 | 2 | 12 |
| IonTorrent | Proton PI | 2012 | 50000000 | 200 | 10 | 4 |
| Illumina (Solexa) | GA | 2006 | 28000000 | 25 | 0.7 | |
| Illumina | GA | 2008 | 28000000 | 35 | 1 | 5 |
| Illumina | GA II | ND | 100000000 | 50 | 5 | |
| Illumina | GAIIx | 2009 | 440000000 | 75 | 33 | 6 |
| Illumina | GAIIx | 2011 | 640000000 | 75 | 48 | 7 |
| Illumina | GAIIx | 2012 | 640000000 | 150 | 95 | 8 |
| Illumina | HiSeq 2000 | 2010 | 2000000000 | 100 | 200 | 9 |
| Illumina | HiSeq 2000 | 2011 | 3000000000 | 100 | 600 | 10 |
| Illumina | HiSeq 2000/2500 | 2014 | 4000000000 | 125 | 1000 | 17 |
| Illumina | HiSeq 2500 RR | 2012 | 600000000 | 150 | 180 | 13 |
| Illumina | HiSeq 2500 RR | 2014 | 600000000 | 250 | 300 | 13 |
| Illumina | HiSeq 4000 | 2015 | 5000000000 | 150 | 1500 | 19 |
| Illumina | HiSeq X | 2014 | 6000000000 | 150 | 1800 | 18 |
| Illumina | NextSeq 500 | 2014 | 400000000 | 150 | 120 | 14 |
| Illumina | MiSeq | 2011 | 30000000 | 150 | 4.5 | |
| Illumina | MiSeq | 2012 | 30000000 | 250 | 8.5 | 11 |
| Illumina | MiSeq | 2013 | 30000000 | 300 | 15 | 14 |
| SOLiD | 1 | 2007 | 40000000 | 25 | 1 | |
| SOLiD | 2 | 2008 | 115000000 | 35 | 4 | |
| SOLiD | 3 | 2009 | 320000000 | 50 | 16 | |
| SOLiD | 4 | 2010 | 2000000000 | 50 | 100 | |
| SOLiD | 5500xl | 2011 | 3000000000 | 60 | 180 | |
| SOLiD | 5500xl W | 2013 | 3000000000 | 75 | 320 | |
| PacBio | RS C1 | 2011 | 432000 | 1300 | 0.540 | |
| PacBio | RS C2 | 2012 | 432000 | 2500 | 1.080 | |
| PacBio | RS C2 XL | 2012 | 432000 | 4300 | 1.858 | |
| PacBio | RS II C2 XL | 2013 | 564000 | 4600 | 2.594 | 15 |
| PacBio | RS II P5 C3 | 2014 | 528000 | 8500 | 4.500 | 15 |
| PacBio | RS II P6 C4 | 2014 | 660000 | 13500 | 9.000 | 15 |
[1] mode or average
[2] Sources: see this file from the github repo.

Very nice and thank you for the update.
For this “certain new instrument”, given your explanation as to why it is *not* included in the first plot, I was wondering if you might not want to also include the “big sibling/army” of this “certain new instrument”, i.e., the PromethION? Albeit with putative numbers of course, something along the lines of https://www.genomeweb.com/sequencing/oxford-nanopore-presents-details-new-high-throughput-sequencer-improvements-mini -> “throughput could increase to more than a terabase per day”.
Best,
Cedric
Thanks for the suggestion, but I’m sticking to my ‘commercially available’ principle here, and will therefore not include the PromethION yet…
On page 10 of the following document you can see the variuos pre-NGS sequencers…
Click to access cms_041003.pdf
The 310 was the smallest with only 1 capillary (1 read/run)
The 3100/3130 had 4 capillaries and the 3100xl/3130xl 16 capilaries (4-16 reads/run)
The 3730 came with 48 capillaries and the 3730xl with 96 (48-96 reads/run)
The capillary length determined the max read length, with the 36cm being ok for 300-400bp, the 50cm hitting around 600bp and the 80cm capillaries pushing over the 1kb
Going further back ABI had the 373 and 377 sequencers which had gel slabs instead of capillaries.. But I never used one so I don’t really know what the specs looked like… But if someone has time to go through the manual here’s a link 🙂
Click to access ABI377manual.pdf
Hope this helps a bit in building the pre-NGS picture
Also, for older instrument the definition of “run” is slightly different… smaller instruments (31xx) can hold 2 x 384-plates whereas the big instruments (37xx) have an autosampler that can hold up to 12 x 384-plates.
If by “run” we mean a single capillary injection (~30min run time) then number of reads/run is equal to the number of capillaries… However if by “run” we mean a session without any hands on time then the throughput goes up significantly. (i.e. 3730xl processing 12 x 384 plates would generate 4608 individual reads and if we assume 800bp we have almost 3,7Mbp output, still tiny compared to NGS but not as low as the picture suggests.
Great great chart nonetheless 🙂
Thanks a lot for the links!
Fantastic – where are you references btw?
Here (as mentioned at the end of the post 🙂 )
What about the Helicos system? Never got the chance to use it but it was kind of the first NGS platforms as well. It might ruin your graph though, since read length is very short and it never evolved…
https://en.wikipedia.org/wiki/Helicos_single_molecule_fluorescent_sequencing
I should add the Helicos, you are right (the thought has crossed my mind). I’ll look into that for the next edition…
It would be incredibly valuable to have some cost metric associated with these data as well. I suspect it would be even more difficult to come up with hard numbers there since it will vary on institution, library prep etc, but there must be some way of consistently calculating it, right?
I agree it would be useful, but I consider it impossibly difficult, for the reasons you mention. It depends heavily on the financing model the different providers have (do users pay just chemicals, or also man hours, amortization, service contracts etc). So I am not going to even try it…
For the PacBio data, do you know if these are these figures based on “raw” (polymerase) reads, or “usable” (sub)reads?
These are raw reads (polymerase reads) metrics. Usuable subread metrics are dependent on the library. I know, it’s not perfect, but not all raw bases from a HiSeq are usable either…
Thanks for the continuous updates. I have been following this since the first edition, looking forward to the next edition.
Btw, specifications for Oxford Nanopore MinION have been released
https://nanoporetech.com/community/specifications
Thanks (had seen that already). I will use that information for the next edition!