I am a fan of the Pacific Biosciences PacBio RS sequencing instrument, as some of the readers of this blog maybe already are aware of. We use data from this instrument in our work, and the Norwegian Sequencing Centre – with which I am affiliated – offers the technology to its users.
PacBio recently increased their read lengths. With this so-called C2XL chemistry, average raw length is now around 4.3-4.5 kbp and maximum read lengths can go over 20kbp. These extra long reads are great for de novo genome sequencing applications, something we are trying ourselves. However, a bit buried in the news about these longer reads is the consequences for PacBio’s so-called Circular Consensus Sequencing, or CCS.
What is CCS?
In order to understand CCS, one needs to know a bit about PacBio sequencing in general. Readers familiar with this may skip to the next section.
The template for PacBio sequencing is a so-called SMRTBell. These are created by ligating hairpin adaptors to both ends of double-stranded DNA molecules. See the figure
The hairpin adaptor has a priming site for sequencing, and the polymerase used for sequencing has strand-displacement capacity (it basically doesn’t care if it has to work through a double-stranded region, it simply ‘kicks-off’ the opposite strand). The effect of this is that the sequence template (SMRTBell) acts like a single-stranded closed circle. The enzyme starts sequencing at the primer location and will sequence the template until it either falls of, or is ‘killed’ as a side-effect of the fluorescent excitation. Given a long enough life for the enzyme, and a short enough insert, the enzyme will in fact go around the hairpin on the other end of the SMRTBell. For the shortest inserts, it can potentially circle around multiple times.

Schematic representation of the SMRTBell template for PacBio sequencing. From http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2926623/
This multi-pass sequencing allows for calling a consensus of the sequence of the insert, overcoming the high single-pass error-rate of the technology (quoted by PacBio as median error rate of ~11%). For successful sequencing in the CCS mode, the insert needs to be short and polymerise life long. A minimum of three passes is needed for the software to even consider calling the consensus, and five or more are preferred to reach 99% accuracy (Q20) or better. One important aspect is the raw read length distribution for PacBio sequencing (see the figure below). The requirement of three or more passes limits the insert size. For example, a 3 kbp raw read is required to get a consensus of a 1 kbp insert. There will therefore always be a – significant! – fraction of raw reads that are not going to yield a consensus read, as they are simply to short. The number of useful reads per SMRTCell (‘chip’) is therefore lower than the number of raw reads useful for long-read (single pass) sequencing.
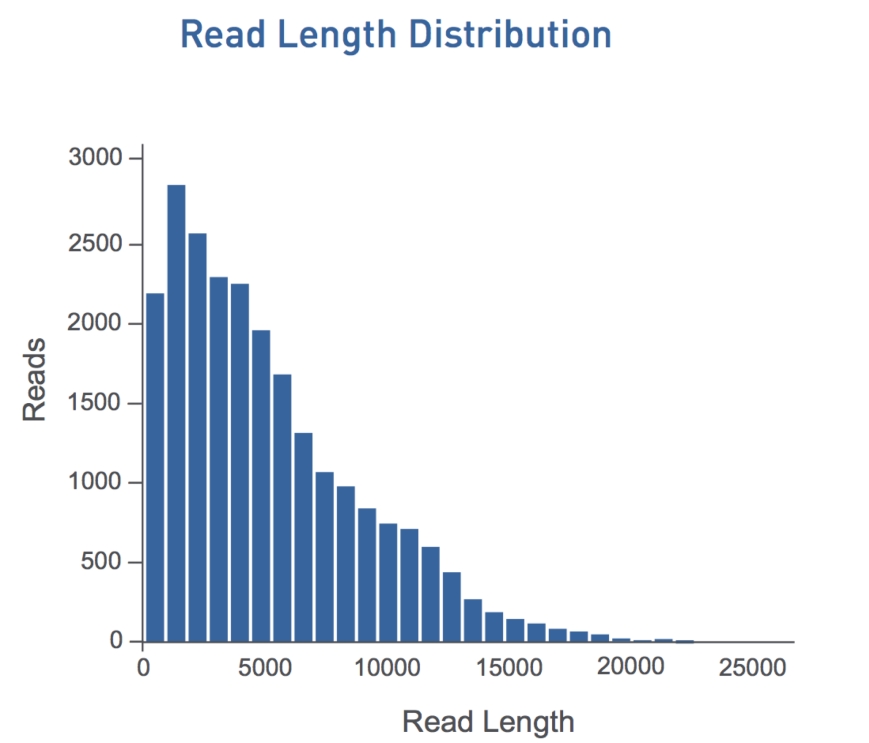
Typical PacBio C2Xl raw read length distribution. From http://pacificbiosciences.com/brochure (February 2013)
CCS and the the new extra-long raw reads
From the above, it follows that an increase in raw read lengths allows for longer insert libraries for CCS reads, or a higher throughput for shorter insert CCS libraries. PacBio used to recommend the 500bp to 1kbp range for CCS libraries. With the C2XL read length increase, this doubles to 1-2 kbp. Throughput is around 40 000 reads per SMRTCell. The remainder of this post describes three possible areas where these extra-long consensus reads may be useful.
Disclaimer
First, I may be a fan of PacBio, and the company is helping us out with our genome projects, but I am not receiving any financial or other personal gains from them. What I write here is my own opinion. Second, the suggestions below are not tested by me or my colleagues – although others may be working along these lines. How the suggested CCS uses I describe work in practice needs therefore to be determined.
1) CCS for sequencing full-length 16S rRNA
Before the days of Next Generation Sequencing, people used to amplify 16S regions of samples and make bacterial clone libraries, sequencing a set of clones using Sanger sequencing. NGS allowed for many more reads per sample, but restricted the length of the sequenced part to a few hundred bases at best. NGS therefore yielded much deeper sequenced datasets, but at a lower discriminatory power (less phylogenetic/taxonomic signal).
The long CCS reads now possible with PacBio imply that one could consider going back to full-length 16S sequencing. The throughput (40 000 reads per SMRTCell) will be much higher than doable using Sanger sequencing, and the quality potentially even better than Sanger. However, the price per read will be significantly higher than using short-read technology. I think that for certain diversity studies, using PacBio CCS with full-length 16S amplicons will be very beneficial.
2) CCS for shotgun metagenomics
Similarly, people interested in whole-sample shotgun metagenomics (as opposed to PCR-based diversity studies) could consider using the 1-2 kbp CCS reads. The long reads could yield much more useful information for gene mining, for example. Some may suggest that using the Roche/454 GS FLX+, which now seems to be working – at least in our lab it does – will yield much more reads (1 to 1.2 million) around that length (we have seen 1kb mode read lengths with GS FLX+!), making this technology more cost-effective. Some back-of-the-envelope calculations using prices for the Norwegian Sequencing Centre show that the comparison actually is in favour of PacBio. Given one library preparation, and 1 million required reads, the 454 is currently out-priced by PacBio, while the latter has the potential to give better quality (no homopolymer errors) and longer reads. However, first, generating 1 million CCS reads (25 SMRTCells) takes more time (several days, not counting library preparation) than a full 454 run (about a day) [Note that lab teams will like not having to do emulsion PCR for PacBio CCS!]. Secondly, the pricing situation may be different for other centres (pricing structures I think really differ from centre to centre).
3) CCS to replace Sanger capillary plate sequencing
Sanger sequencing is far from dead. Its main attraction is that it allows very small sample sizes (down to a single sample can be submitted to a facility), and long reads with high quality. A key difference between Sanger and NGS is the fact that each read can be traced back to a well on a plate. I think that, if a suitable and cost-effective barcoding scheme could be designed for multiplexed PacBio CCS sequencing, PacBio CCS could potentially replace Sanger plate sequencing. To keep the costs down, one would need to massively multiplex, perhaps dozens of 96 well plates with fragments that need to be tracked back to their original plate and well. But a laboratory with good automation experience might pull it off. At the same time, the scheme requires a steady flow of Sanger samples. It wouldn’t work for a facility that sequences less then, say, a dozen plates per week. Commercial providers may actually already consider doing this switch. The benefits could be longer reads than one can get with Sanger, with higher per-base qualities.
[Technical note: the per-SMRTCell throughput of 40 000 reads may allow for adding the same template multiple times, increasing the final consensus accuracy. The fraction of raw reads too short to give a consensus call (see above) may actually contribute to quality as well as they are barcoded]
In summary: PacBio CCS may be an alternative for short read sequencing, or even Sanger Capillary sequencing. However, there will be a trade-off in information content, versus price per read.
For a technical, but very readable paper describing CCS (from the PacBio researchers themselves), see this paper in Nucleic Acids Research.

Hi,
We had PacBio sequencing data constructed with CCS library. After some preliminary analysis, I found only 7% of all the reads has greater than 3 passes and yield a collapsed CCS read. The rest of them only has 1~2 passes or plus some incomplete pass. So my question is, is this normal in pacbio data? If so, how can I analyze it. Because blasr align the linear sequence to multiple random location on the genome for each pass segment with high variability. And meanwhile, is there a way to assemble the sequence from a CCS library since I read the paper suggesting that for CLR reads only.
Best,
Jack
I don’t know what would be a “normal” distribution of the number of passes – it also depends on the insert size and raw sequencing length. Have a look at this recent paper: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3811116/. I don’t know why you get random (i.e., non-overlapping?) placement of each subread, perhaps they are from a repeated area? If you add the “-useccs” flag, blasr aligns “the ccs sequence, then report[s] alignments of the ccs subreads to the window that the ccs was mapped to. Only alignments of the subreads are reported.” I wouldn’t know about assembly – the paper I mention describes a brief pipeline. There are not that many programs that can tackle long reads, but MIRA and Celera can.