This is the third edition of this visualisation, previous editions were in October 2013 and December 2012.
As before, full run throughput in gigabases (billion bases) is plotted against single-end read length for the different sequencing platforms, both on a log scale:
A new visualisation
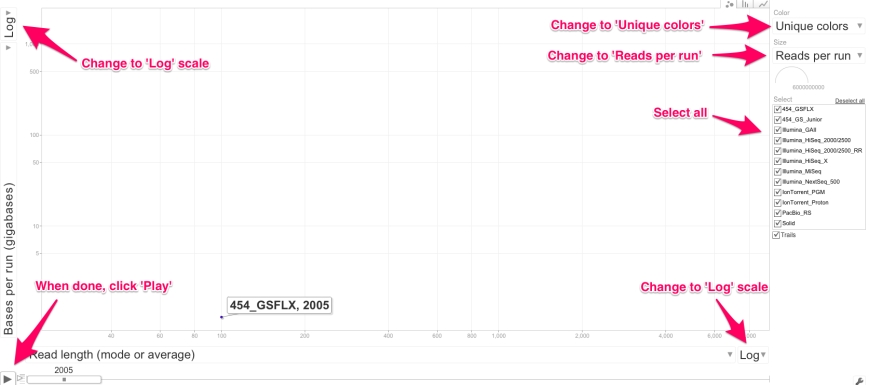
Inspired by GapMinderWorld, a fascinating interactive visualisation of demographic data, and using their recommendations, I created an interactive ‘Motion Chart’ version of this visualisation on a Google spreadsheet. The chart allows to track the metrics throughout the years. Here is the final graph after running though all the data:
The X and Y axis are the same as for the first figure, the size of the data points are correlated with the number of reads per run.
You can explore the data interactively yourself by clicking on the graph.
Unfortunately, I didn’t find a way to change the default chart settings, so in order to have the best experience, make sure to adjust the chart as depicted below:
Note that the log scaling does not work as well as in the static picture at the top of this post. This is the reason I did not include the datapoint for Sanger sequencing. If someone can make this graph work in, say python, I’d be happy to include your results!
Notable changes from the October 2013 edition
- I use numbers for the full run output. It was pointed out to me that this was not the case for the PacBio data, where I so far used the metrics for single SMRTCells (‘chips’) only. I have now chosen to report metrics for 12 SMRTCells as a full PacBio run, a compromise between the 8, 12 or 16 SMRTCells per run we have worked with
- PacBio also upgraded to P5-C3 metrics
- the Roche/454 GS Junior upgraded the read length to 700 bp (‘GS Junior+’)
- the Illumina HiSeq2500 ‘1TB’ upgrade (2 × 125 bp read length and 4 billion reads per run)
- I added the Illumina NextSeq 500 and HiSeq X (I chose the output for 1 instrument, even though one has to buy at least 10 of them)
Some comments
- note how close together the data points fall for the GAII, HiSeq ‘Rapid Run’ mode and NextSeq 500.
- as mentioned in the original blog post: some data was obtained by going to previous versions of company websites through the Internet Archive
- I used full single-run specs with maximally stated throughput as available at the time of writing
- sometimes, the total numbers of reads per full run and total bases obtained do not match up; for the figure, I always chose the reported throughput in bases
- for Illumina, I chose to use the single-end read length, although the maximum throughput was based on the sum of all reads from a paired end run; I felt it unfair to double the read length for this platform for the figure
- no changes for the 454 GS FLX+, Illumina GAII, HISeq ‘Rapid Run’ mode, SOLiD, Ion Torrent PGM and Proton
- Oxford Nanopore’s MinION was not added as the instrument is not yet full commercially available – they are still in early access phase (MinION Access Program)
Availability
Data and figures are released under a CC0 license at figshare, with doi 10.6084/m9.figshare.100940. I’ve also added the content to Github at https://github.com/lexnederbragt/developments-in-next-generation-sequencing.
Disclaimer
As before: although I took utmost care in collecting the data, I may have gotten some of my numbers completely wrong, for which I apologise in advance; please help me correct any mistakes or omissions through leaving a comment, or sending me a pull request.
Finally, the raw data
| Platform | Instrument | Year | Reads per run | Read length[1] | Gigabases per run | Source [2] |
|---|---|---|---|---|---|---|
| ABI Sanger | 3730xl | ND | 96 | 800 | 0.0000768 | |
| 454 | GS20 | 2005 | 200000 | 100 | 0.02 | |
| 454 | GS FLX | 2007 | 400000 | 250 | 0.1 | |
| 454 | GS FLX Titanium | 2009 | 1000000 | 500 | 0.45 | |
| 454 | GS FLX+ | 2011 | 1000000 | 700 | 0.7 | 1 |
| 454 | GS Junior | 2010 | 100000 | 400 | 0.04 | 2 |
| 454 | GS Junior+ | 2014 | 100000 | 700 | 0.07 | 16 |
| IonTorrent | PGM 314 chip | 2011 | 100000 | 100 | 0.01 | 3 |
| IonTorrent | PGM 316 chip | 2011 | 1000000 | 100 | 0.1 | 3 |
| IonTorrent | PGM 318 chip | 2011 | 5000000 | 100 | 0.5 | 3 |
| IonTorrent | PGM 318 chip | 2012 | 5000000 | 200 | 1 | 3 |
| IonTorrent | PGM 318 chip V2 | 2013 | 5000000 | 400 | 2 | 12 |
| IonTorrent | Proton PI | 2012 | 50000000 | 200 | 10 | 4 |
| Illumina | GA (launch?) | 2006 | 28000000 | 25 | 0.7 | |
| Illumina | GA | 2008 | 28000000 | 35 | 1 | 5 |
| Illumina | GA II | ND | 100000000 | 50 | 5 | |
| Illumina | GAIIx | 2009 | 440000000 | 75 | 33 | 6 |
| Illumina | GAIIx | 2011 | 640000000 | 75 | 48 | 7 |
| Illumina | GAIIx | 2012 | 640000000 | 150 | 95 | 8 |
| Illumina | HiSeq 2000 | 2010 | 2000000000 | 100 | 200 | 9 |
| Illumina | HiSeq 2000 | 2011 | 3000000000 | 100 | 600 | 10 |
| Illumina | HiSeq 2000/2500 | 2014 | 4000000000 | 125 | 1000 | 17 |
| Illumina | HiSeq 2500 RR | 2012 | 600000000 | 150 | 180 | 13 |
| Illumina | HiSeq X | 2014 | 6000000000 | 150 | 1800 | 18 |
| Illumina | NextSeq 500 | 2014 | 400000000 | 150 | 120 | 14 |
| Illumina | MiSeq | 2011 | 30000000 | 150 | 4.5 | |
| Illumina | MiSeq | 2012 | 30000000 | 250 | 8.5 | 11 |
| Illumina | MiSeq | 2013 | 30000000 | 300 | 15 | 14 |
| SOLiD | 1 | 2007 | 40000000 | 25 | 1 | |
| SOLiD | 2 | 2008 | 115000000 | 35 | 4 | |
| SOLiD | 3 | 2009 | 320000000 | 50 | 16 | |
| SOLiD | 4 | 2010 | 2000000000 | 50 | 100 | |
| SOLiD | 5500xl | 2011 | 3000000000 | 60 | 180 | |
| SOLiD | 5500xl W | 2013 | 3000000000 | 75 | 320 | |
| PacBio | RS C1 | 2011 | 36000 | 1300 | 0.540 | |
| PacBio | RS C2 | 2012 | 36000 | 2500 | 1.080 | |
| PacBio | RS C2 XL | 2012 | 36000 | 4300 | 1.858 | |
| PacBio | RS II C2 XL | 2013 | 47000 | 4600 | 2.594 | 15 |
| PacBio | RS II P5 C3 | 2014 | 44000 | 8500 | 4.500 | 15 |
[1] mode or average
[2] Sources: see this file from the github repo.



Hi Lex,
thank you for the nice graph! I will include it in my thesis.
Also the DOI hyperlink in the “Availability” section is incorrect.
Cheers,
Dave
Thanks! The doi/dead link should be fixed now.
Hi,
Thanks for the updated table. You are correct, 2006 is launching, but it should show Solexa in the platform, as Illumina bought it in 2007: http://www.sciencedirect.com/science/article/pii/S1871678409000089
I think the nature paper refers to the two paper about the technology evolving and acquisition:
http://www.nature.com/nbt/journal/v26/n10/full/nbt1486.html#B22
Thanks for confirming the launch year. Technically you are correct that in 2006 the company name should be Solexa. This will be addressed in the next edition. Thanks!
Too bad you used the worst of the Sanger instruments for your comparison, but the difference is still stark.
The Amersham/GE Healthcare MegaBACE 4500 was a 384-capillary instrument, with readlengths over 1000 bp. Yet, due to ABI’s better dominance in the market, the MB 4500 never had much penetration.
Disclosure: I developed the MegaBACE 4500.
Hmm, you’re giving me a dilemma: yes, I could add other then-available instruments, which would be nothing else than fair. however, I have no overview of which these were, when they were available, and what the relevant metrics were. Or, if I don’t, I should remove the ABI… I’ll give this a bit of thought. Oh, and when I just started working here in 2005, we still had a MegaBACE – although I never used it.
Hi, could I use one of your illustration in my master thesis? i would indicate the source obviously..
thanks in advance
Absolutely!
Hi. Excellent summary which I have shared around my institution. As a PGM evangelist can I point out that 314v2 and 316v2 chips were launched a while back enabling 400bp reads on those chips. Also I presume you’re comparing on an “even” grounding and using the official manufacturer’s numbers? I only mention as I would be pretty disappointed with a 314v2 run that didn’t come out with >250,000 reads and 316v2 that didn’t come to >3m
Thanks! My bad for not following along. Was this release in 2014 or 2013? I need to update the graph (HiSeq4000! RapidRun v2! pacBio P6-C4!) so then I will add your suggested updates.
The v2 chips and 400 bp kit came out in 2013. Thanks again for your efforts.
I only now see you are talking about the v2 for the 314 and 316 chips. I decided to take the maximum output chip for the PGM, i.e.the 318, to not clutter the graph with too many data points and lines for this instrument.
Dear Lex,
Just one more historical update to your’s table
The ABI 3730/3730XL was released in summer 2002 to commercial users (see this link):
http://www.eurekalert.org/pub_releases/2002-04/pn-abi042302.php
Before it there was ABI 3700 96cap (released in 1998), which had a bit shorter read length (Q20 was arround ~550bp or so), but it depended on array lenght (50cm) run voltage/time, polymer used, etc:
http://www.medwow.com/med/genetic-analysis-system/applied-biosystems/abi-prism-3700-genetic-analyzer/39115.model-spec
Thank you! Such information is not easy to find (I’m still uncertain about release dates for SOLiD…). I will try to incorporate this information for the next release (it’s about time for an update, really…)
Great post, fantastic graphics!
Is there a 2016/2017 version?
There will be, as soon as I find time to make it…
Pingback: Difference Between Microarray and Next Generation Sequencing – In4arts.com