PacBio sequencing is all about looong reads, especially in relation to de novo sequencing. A few things are needed to get the longest reads possible:
- the longer the enzyme is active on the template, the longer the raw read will be – PacBio calls these ‘Polymerase reads’
- a library for pacBio sequencing consist of circular molecules, with the target insert between two hairpin adaptors, allowing the enzyme to ‘go around’ and sequence the opposite strand once it reaches the end of the insert. See my previous post on this here. It then follows that the longer the template used for library preparation, the smaller the chance the polymerase goes around the hairpin, leading to longer uniquely sequenced template – PacBio calles these ‘reads of insert’ – and these represent the most useful reads for de novo sequencing applications
- finally, the distribution of sizes of the library has an influence: any high-throughput sequencing technology, as well as PCR, has problems with ‘preferential treatment’ of smaller fragments. With PacBio sequencing, shorter molecules tend to load preferentially into the wells of the SMRTCell (‘chip’). It then makes sense to try to reduce the shoulder of shorter fragments for the final library preparation.
Recently, PacBio and Sage Science announced a co-marketing partnership for the BluePippin. This instrument allows for tight size selection of DNA samples, effectively making the peak of the size distribution much more narrow. With regard to PacBio sequencing, a narrow peak lessens the problem of preferential loading of short fragments, leading to much longer ‘reads of insert’. A demonstration can be seen on this poster (I think I know which fish they used for that one plot…).
It’s nice when a company demonstrates a new improvement to their technology. But, the proof is always in the pudding, in this case, can the sequencing centres out there actually show the same results? Based on recent tweets from the PacBio USA User group meeting, one would believe so:
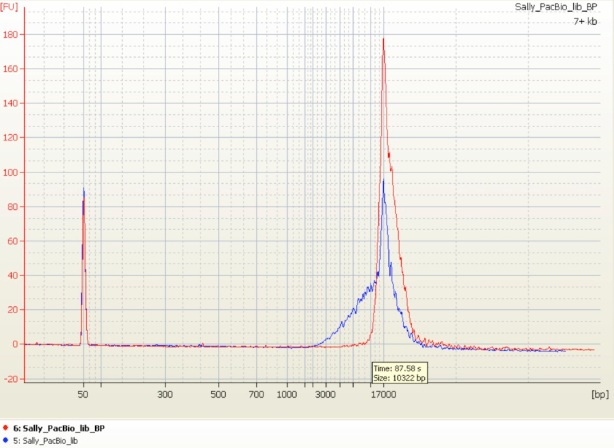
The Norwegian Sequencing Centre, with which I am affiliated, recently bought a BluePippin instrument for the reasons outlined above. Our first test was on a good, 10 kb insert PacBio (SMRTBell) library that we already had used for sequencing. The (excellent!) lab team ran a (large) sample of the library on the BluePippin instrument and showed a significant reduction of the small fragments:
Note that we didn’t try to remove the larger fragments (which you would probably do for mate pair libraries) because in the case of PacBio library preparation, length is more important than a tight distribution per se. The next question was what the effect is of such a cleanup has on sequencing. We ran both libraries on our PacBio instrument with C2XL chemistry and 120 minute movies. First some numbers. Please note that the regular library data was obtained from our PacBio RS before maintenance and upgrade to RSII (doubling of the laser capacity), and this may have had a lsight negative effect on the read lengths. There were also more reads included for the ‘before’ data, and these were analysed with version 1.4 of the PacBio software versus 2.0 for the BluePippin library.
| Library | # of reads | Ave. length | Longest subread N50* | Longest |
| Regular library | 693,275 | 3,009 bp | 4,041 bp | 22,298 bp |
| BluePippin library | 325,407 | 6,045 bp | 8,820 bp | 25,931 bp |
*N50 is a metric often used for genome assemblies, and here translates to ‘the length at which half the bases of the entire dataset is in reads of at least length N50′.
The following graph shows the distribution of the longest subreads (or, reads of insert) for each raw read produced, for the library before, and after BluePippin cleanup.

Length distribution of longest subreads from the sequenced libraires before and after BluePippin cleanup
The plot shows how the length distribution shifts significantly to longer reads after BluePippin cleanup. The peak at very short readlengths for the regular library is I think an effect of a missing ‘minimum subread length’ filter for splitting of these reads.
A better way to visualise this is with a plot that shows the fraction of bases at different length cutoffs:

Fraction of bases at different length cutoffs of longest subreads from the sequenced libraries before and after BluePippin cleanup
The stippled lines show the N50 length. The plot shows that the BluePippin prep indeed had the desired effect: the reads are much longer.
These very long reads are ideal for de novo assembly, provided that they can be error-corrected. PacBio, as well as a number of research group, have demonstrated that error-correction, using high-quality short reads, or high-coverage PacBio single-pass reads, results in long, high-quality reads that can yield (close to) finished bacterial genomes and improved larger genomes.
A drawback of involving the BluePippin is that more DNA may be needed for library preparation. Due to the fact that PacBio sequencing is single molecule, without amplification of the library before sequencing, there usually is a limited number of SMRTCells one can obtain from a library. A lot of DNA is lost during library prep, and also with the BluePippin.
A final note: to be able to obtain average subread lengths (‘reads of insert’) that high (over 6kb), the raw read (‘polymerase read’) length average also needs to be at least that high. And indeed, for some reason we don’t know yet, the BluePippin library showed much larger average raw readlengths than a regular library. The BluePippin library showed an average readlength between 5.5 and 6.5 kbp, while the current specifications mention 4.6 kbp. We have already seen an effect of organism sequenced on the raw read length. All of this is probably due to the fact that PacBio sequencing is single-molecule sequencing, allowing for species (or sample)-specific properties to affect polymerase speed. We have now started sequencing bacterial genomes with BluePippin treated libraries, and those show even longer average raw readlengths (more on that perhaps in a later post).
Thanks to the Norwegian Sequencing Centre lab team who made this post possible.
Code used
The code is available as an iPython notebook viewable here, and downloadable here.

Nice analysis. In future plots, could you assist the color-challenged by also differentiating your curves by line type (dotted vs. solid) or weight?
Good tip! Having the usual male colourblindness problem myself I can relate…
Working with pacbio team also used blue pippin for size selection got the same result. It will be nice blue pippin give us better yield,it eats most of the DNA.