(The impatient reader might want to skip to the conclusion at the end of this post…)
Last wednesday, Ion Torrent released a tech note and associated run data with shotgun (single-end) and Mate Pair runs for Escherichia coli K12, substrain MG1655. Both a 3.5 kb and 8.9 kbp insert size, as well as a shotgun library, were sequenced on a 316 chip each. In the tech note, they describe assemblies using different combinations of the data, and show how adding the mate pairs yields assemblies with fewer scaffolds and gaps. The Ion mate-pair protocol is very similar to the one used by 454 Life Sciences for their (unfortunately called) Paired-end libraries: long fragments are circularized using a linker sequence, and sequencing is peformed across this linker, allowing for easy identification of the pair halves.
This is the first real ‘long-distance’ Mate Pair data from Ion Torrent, which is exciting and made me have a close look at it. I was especially interested in how the newbler program, developed by 454 Life Science for their 454 reads, would perform on these data.
Mapping
First, some impressive numbers: the mate pair runs yielded 2.5 – 3 million reads (average lengths 192 and 200 bp). When you generate these kinds of libraries for bacterial genomes using 454, one would ordinarily sequence 1 lane of a plate divided into 8 lanes, and get less then 1 tenth of the throughput.
I first mapped the mate pair runs to the reference genome sequence using gsMapper from the newbler software suite. I immediately came across the first problem: the linker sequence used by Ion is different from the one used by 454. This means that newbler does not recognize the reads as mate pairs. I am really hoping a future version of newbler will allow for custom mate pair linker sequences, but given that Ion Torrent is a big competitor for Roche/454, I doubt this will ever happen…
So, I resorted to what Ion also used: taking the sff_extract program to split the mate pair reads, and outputting them as fasta and qual files. Examining the results showed that 37% of the reads from the 3.5kb run were mate pairs (had the linker according to sff_extract), and 25% for the 8.9 kbp run. The remainder of the reads were shotgun reads. As for the 454 mate pair protocol, there is always going to be a fraction of non-linker-containing reads. However, the fractions for the Ion runs are considerably higher than what we usually get for 454 mate pair (‘paired end’) runs. However, the higher throughput with Ion Torrent more than compensates for the low fraction of mates.
After extracting the pair halves, I adjusted the files so that newbler would accept them as mate pairs, and performed mapping using newbler (gsMapping/runMapping program). Then I plotted the distances between the mapped mate halves for both libraries:
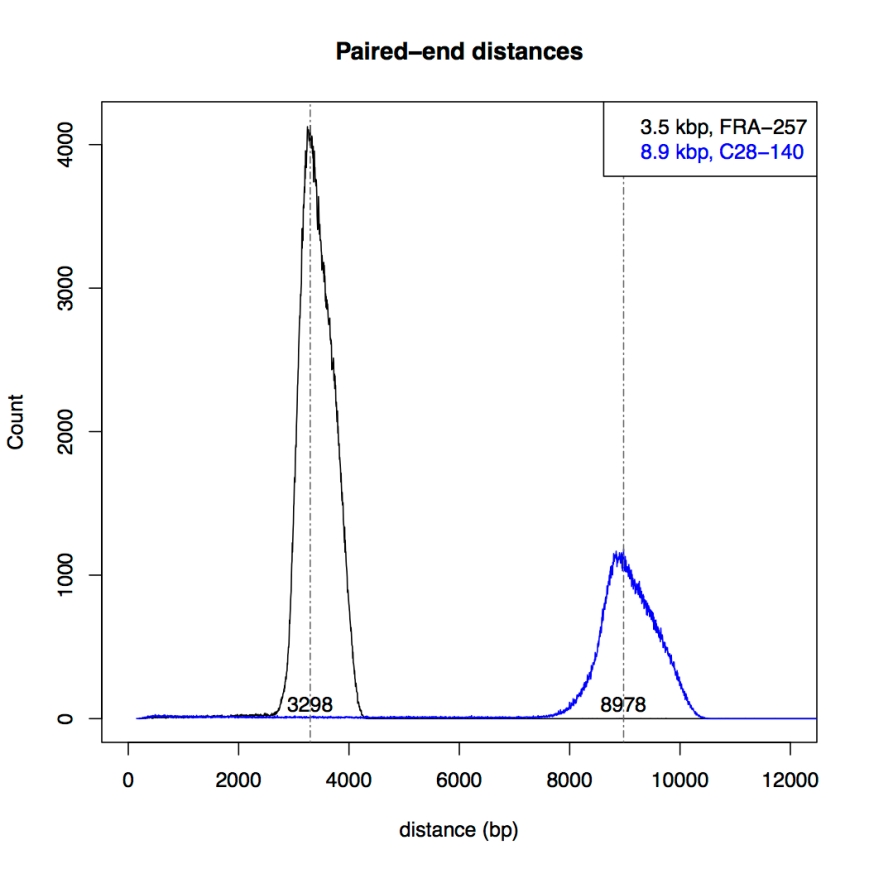
The figure shows a nice distribution with sharp peaks around 3.3kb and 9kb, respectively, confirming the mapping shown in Ion Torrents tech note.
Assembly
Next, I set out to perform assemblies. Ion Torrent provided files with reduced amounts of reads so that there would be about 20X and 40X coverage of the 5.4 Mbp genome. I decided to use these files, rather than make my own scaled down versions. I do note, however, that the 20X and 40X files of the mate pair library runs mostly contain mate pair reads, and very few singletons (based on the sff_extract results). Whether Ion determined which reads were mate pair reads by mapping to the reference genome, or by using sff_extract, and only taking reads that were split along the linker, I cannot tell (in a real de novo setting, the second option is the only possible, of course).
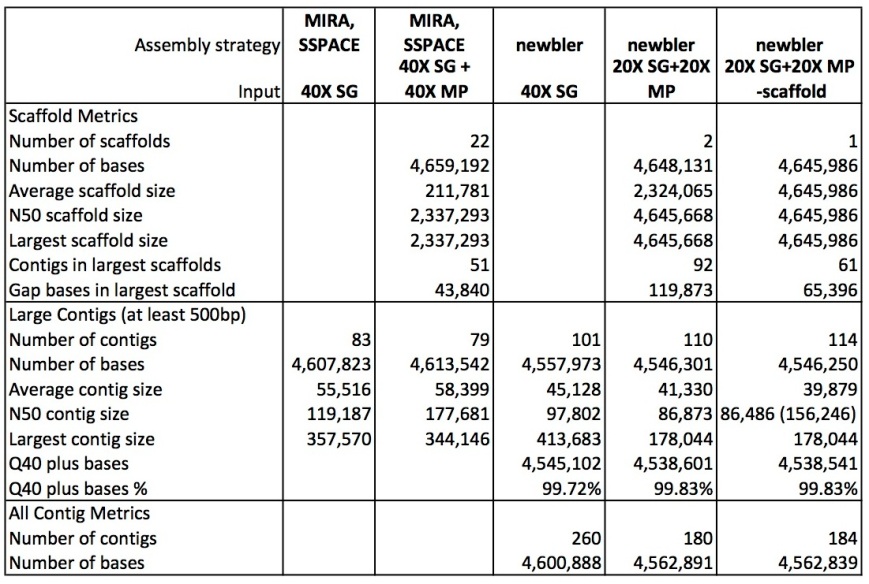
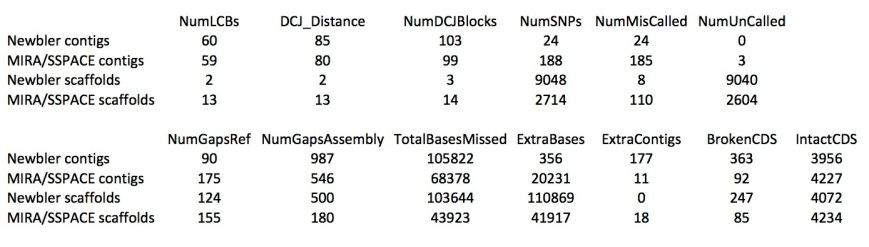
The table below shows metrics for five assemblies. The first two are described in the Ion Torrent tech note as being done using MIRA for contig building and SSPACE for scaffolding:
– the first column is for the shotgun (single-end, fragment reads) only contigs
– the second column for the scaffold.
Column 3 to 5 describe the three assemblies I produced for this post using newbler:
– using the shotgun reads only (40X coverage)
– using shotgun (20X) plus the 3.5 kb and 8.9 kb mate pair reads (20X each)
– using shotgun (20X) plus the 3.5 kb and 8.9 kb mate pair reads (20X each) but with the addition of the ‘scaffold’ option; newbler here fills gaps caused by repeats with a consensus sequence of the repeat copies
Newbler manages to generate a single scaffold for the this genome! (the second scaffold in the ’20X SG+20X MP’ assmbly is a contig larger than 2 kb, which newbler automatically lists under the scaffolds). However, the contigs produced by MIRA are longer than those produced by newbler. Also, MIRA assembled the genome into fewer contigs compared to newbler, and with less gap bases. Looking closer at the MIRA/SSPACE scaffolds shows that the genome is in fact assembled in four large pieces of 2.3 Mbp, 1.3 Mbp, 700 kbp and 200 kbp, with another 10 kbp scaffold (rRNA?) and 17 very small scaffolds.
I tried different assembly strategies, but I could not improve on the N50 and longest size reported here. By the way, the N50 contig size in brackets is for the scaffolded contigs in the ‘-scaffold’ assembly, where gaps have been filled using consensus repeat sequences, thereby merging contigs into single, bigger ones.
Reducing the coverage to 20X shotgun + 10X mate pair (each library), or even 20X shotgun and only the 8.9 kbp mate pair reads also resulted in a single scaffold for the genome (‘not shown’).
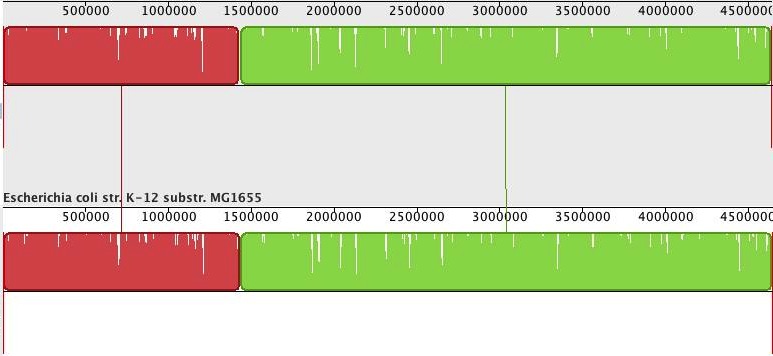
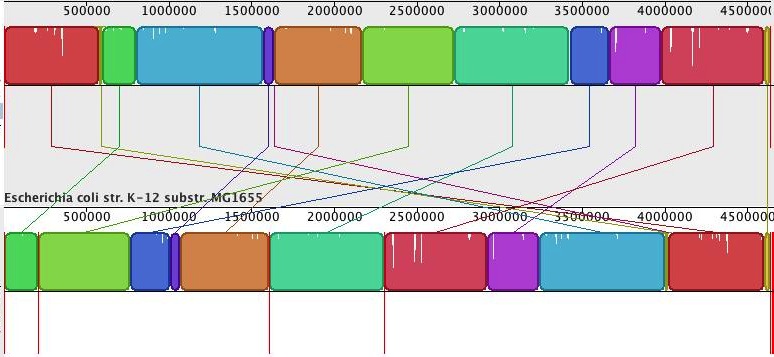
Now, it is nice to have ‘better’ assemblies in terms of contig or scaffold lengths, but how good are these, really? Luckily, we can compare the assemblies to the reference genome. For this, I used Mauve Assembly Metrics (see also Nick Loman’s post about this program here). (Due to a bug in the Mauve, at least that is what I think caused the crash, I could not get the ‘-scaffold’ newbler assembly analysed). The program gives a lot of results, but I will only share the most important ones here. First, the alignment of the scaffolds against the reference (scaffolds reordered relative to the reference genome for the Ion Torrent assembly). Newbler scaffolds:
MIRA/SSPACE (done by Ion Torrent) scaffolds:
Whereas the alignment is perfect for the single newbler scaffold, the MIRA/SSPACE scaffolds show numerous misassemblies (vertical red bars depict scaffold boundaries).
Some numbers from Mauve:
SNPs seems to be the sum of uncalled bases (‘N’s) and miscalled bases. Newbler seems to be able to assemble with fewer miscalled bases, but with many more uncalled bases (although Mauve reports far fewer than the assembly does). I don’t really understand the difference between gaps in the reference and in the assembly. MIRA misses fewer bases (gaps), but it’s contigs (not it’s scaffolds) have more extra bases (insertions). Also, the set of newbler contigs contains some few (probably very small) contigs that could not be mapped to the reference genome. Finally, the number of intact and broken CDSs (coding sequences of genes) numbers don’t surprise me, as the contigs of MIRA are so much longer, they are able to capture much more of the gene space. MIRA contigs already have 98% of the CDSs complete.
Conclusions
Ion Torrent seems to have done a good job in enabling mate pair sequencing on their platform, with nice tight size distributions, and impressive throughput relative to 454. These mate pair reads, together with the shotgun (single end) reads, are very useful for de novo assembly. The de novo assembly approach Ion Torrent chose, using sff_extract, MIRA and SSPACE, seems to be giving quite long contigs, with almost all genes complete. However, newbler outperfoms SSPACE in scaffolding. Newbler is able to assemble the reads into a single scaffold, even with shotgun reads only supplemented with the 8.9 kb mate pairs. However, newbler’s algorithm is not able to produce as long contigs as MIRA does. So, a viable strategy for de novo assembly, using Ion Torrent shotgun (single end) plus mate pair reads, would be to generate contigs with MIRA, contigs and scaffolds with newbler, and elongate the newbler contigs with the MIRA contigs to reduce the number of gaps in the newbler scaffold(s). What is missing from this analysis, though, is a comparison of a similar assembly using 454 reads, something I hope to be able to do in the near future…
With adding the possibility of long insert mate pair library construction and sequencing, Ion Torrent has moved into another niche of Roche/454 Life Sciences. With the fast run time and high throughput of the PGM relative to the 454, Ion Torrent is on its way to become a viable option for de novo small genome sequencing. Apparently, making longer insert libraries (> 10kb) is cumbersome using the Illumina protocols, but not so much with the 454 version, and hopefully also feasible with the Ion Torrent protocol. In addition, the use of a linker allows for easy separation of the mate halves relative to the linker-less Illumina mate-pair reads. It remains to be seen, though, if research groups are able to generate these mate pairs independent of the Ion Torrent labs.
Code Used
(As always, let me know in the comments if things are unclear of missing, or didn’t work out for you.)
Data and programs
I downloaded some of the sequencing run files mentioned in the tech note, and unzipped them. For the ‘whole run’ files, I needed a newer version of unzip than the default installed on our system. So, I downloaded that from http://www.info-zip.org/. I also downloaded the MIRA assemblies, the linker sequence file and the reference sequence, but for the annotated version (for Mauve), I went to Genbank instead.
I download sff_extract 0.2.13 and, since it needs this program to find the linkers, ssaha 2.5.5. I also installed Mauve 2.3.1 and Mauve assembly metrics scripts.
Preparing the reads
For getting the mate pairs split along the linker, I used (for runs FRA-257 and C28-140)
sff_extract_0_2_13 -c -l LMP_Linkers.fasta somefile.sff
The ‘-c’ option ensures the clipping points present in the sff file are used to give trimmed sequences. This takes quite some time and uses a lot of space on /tmp (I could only run one sff_extract at a time). This gives three files: a fasta file, a corresponding qual file, and an xml file which I didn’t need (but MIRA, for example, needs it).
The sequence headers of the resulting fasta files look like this:
>PT1DY:2303:781.r
>PT1DY:2303:781.f
(for the forward and reverse half, respectively).
In order for newbler to accept these reads as paired (mate pairs), they need to look like this:
>PT1DY:2303:781.r template=PT1DY:2303:781 dir=r library=FRA-257_3.5kb
>PT1DY:2303:781.f template=PT1DY:2303:781 dir=f library=FRA-257_3.5kb
I used the following command to change the headers:
cat FRA-257_3.5kb.fasta |awk '{if (!/>/){print $0}
else{split($1,e,"."); print $1" template="substr(e[1],2)" dir="e[2]" library=FRA-257_3.5kb"
}}' >FRA-257_40X_for_newbler.fasta
Same for the corresponding ‘qual’ file. Important: in order for newbler to incorporate the quality vaules, the qual file needs to have the same name, with ‘fasta’ replaced by ‘qual’, so in this case ‘FRA-257_40X_for_newbler.qual’
I adjusted the ‘library=’ accordingly for the 8.9 kbp mate pair reads.
Read statistics
In order to get the statistics for the percentage of mate pair reads, I used the sff_extract output. this program will annotate reads as being ‘f’ and ‘r’, mate par halves, ‘fn’, no-mate pair read (regular shotgun read), and ‘part#’, when only a partial linker was found. As an example, for the full run for the 3.5 kbp library, FRA-257, I used these commands.
To summarize the different sff_extract ‘categories’ (with output from the command):
cat FRA-257.fasta |awk '/>/ {split($1,a,"."); x[a[2]]++}END{for(i in x){print i"\t"x[i]}}'
fn 1557426
f 880617
r 880617
part1 23495
part2 22337
part3 5
To get the total number of unique reads:
cat FRA-257.fasta |awk '/>/ {split($1,a,"."); x[a[1]]++}END{for(i in x){print i"\t"x[i]}}' |wc -l
2461538
So, 1557426 ‘fn’ reads out of 2461538 = 63%, leaving 37% of reads being mate pairs.
Mapping
Using newbler 2.6, and with the provided reference file (MG1655.fasta), I ran:
runMapping -o map_all_matepair MG1655.fasta FRA-257_for_newbler.fasta C28-140_for_newbler.fasta
(newbler will recoginze the ‘.qual’ file if has the same name, and is located in the same folder as the ‘.fasta’ file).
For plotting the insert distances, I used the following R script:
# extract from the 454PairStatus.txt file:
# - the first 5 characters from the read name
# - the pair-halves distances
d = read.table(pipe("cat 454PairStatus.txt | awk '$3>0{print substr($1,1,5),$3}'"), header=T)
# 3.5 kb library
h3=hist(d[d$Templ=="PT1DY",2],breaks=0.25*max(d$Distance),plot=F)
peak3=h3$mids[which(h3$counts==max(h3$counts))]
# 8.9 kb
h9=hist(d[d$Templ=="6S604",2],breaks=0.25*max(d$Distance),plot=F)
peak9=h9$mids[which(h9$counts==max(h9$counts))]
pdf("AllPairDist.pdf")
plot(h3$mids,h3$counts,type="l",xlim=c(0,12000),main="Mapped paired-end distances",xlab="distance (bp)", ylab="Count")
lines(h9$mids,h9$counts, col="blue")
abline(v=peak3,lty=6,col=gray(0.4))
text(peak3,100,labels=round(peak3))
abline(v=peak9,lty=6,col=gray(0.4))
text(peak9,100,labels=round(peak9))
legend(x="topright", legend=c("3.5 kbp, FRA-257","8.9 kbp, C28-140"),text.col=c("black","blue"),bg=gray(1))
dev.off()
Note how, instead of reading in the entire 454PairStatus.txt file, I use the ‘pipe’ trick of R to get the output of a shell command instead…
Assemblies
Using again newbler 2.6, I ran, for instance:
runAssembly -o 20xSG+20xMP -p FRA-257_20X_for_newbler.fasta -p C28-140_20X_for_newbler.fasta C11-127_20X.sff
The ‘-p’ flag forces newbler to trat the files as mate pairs (paired end, in newbler technology). I used the native sff file for the shotgun run.
Assembly statistics were obtained using my newblermetrics script, MIRA assembly metrics were from the tech note.
Mauve
I first collected the genbank reference file, and relevant assemblies (either contigs, or scaffolds) in a single folder, but rather then copying the files, I made symlinks to them. Then, I used a shell script to run this programn, as per the website:
export PATH=$PATH:/path/to/mauve_snapshot_2011-07-18
export CLASSPATH="/path/to/mauve_snapshot_2011-07-18/ext/*"
# run the scoring
java -cp /path/to/mauve_snapshot_2011-07-18/Mauve.jar org.gel.mauve.assembly.ScoreAssembly -reference NC_000913.gbk -assembly Ion.fa -reorder Ion -outputDir Ion
java -cp /path/to/mauve_snapshot_2011-07-18/Mauve.jar org.gel.mauve.assembly.ScoreAssembly -reference NC_000913.gbk -assembly newbler.fa -reorder newbler -outputDir newbler
# Generate plots of metrics
# ./ indicates the parent directory containing the assembly scoring directories -- the current working directory in our example
mauveAssemblyMetrics.pl ./
I opened the ‘alignmentx’ file in mauve from the ‘alignmentx’ folder with the highest number. The table was based on the ‘summaries.txt’ files produced.

Thanks for this description of working with mate pair data. Do I understand correctly that you are dealing with 2-3 million 200 bp reads each as an individual fasta file? Does this place any strain on the filesystem you’re using?
The biggest sff file from this dataset was C11-127.sff, and it clocked in at 5 Gb. This is not extraordinary, especially when you consider Illumina sequence file, which can be this size after compression… Sure, things get slower with these large files, but one just has to adapt…
Hello!
Just a quick question…
When they talk about “3.5 kbp and 8.9 kbp insert size”, how exactly is this distance measured relatively to the reads?
– from the *first* base of the 1st read to the *first* base of the 2nd read (difference between starting positions) ?
– from the *last* base of the 1st read to the *first* base of the 2nd read (space between the two mates) ?
– from the *first* base of the 1st read to the *last* base of the 2nd read (full size including the two mates) ?
And do you know if this distance is calculated the same way in 454 and in Illumina?
Thank you very much for your attention!
How it is calculated depends not on the technology (454 vs ion), but on the program. Newbler, as far as I know, uses “from the *first* base of the 1st read to the *last* base of the 2nd read (full size including the two mates)”. I’m unsure about other programs, but it seems logical they use the same – this represents the length of the original fragment used for circularization.
Thank you very much for your reply!
Yes that is also the definition in the SAM format specification (Section 1.4.9: TLEN).
But actually I was not referring to the way it is *calculated* by software but more to the way it is *generated*.
After some further research, I found out that both Illumina and IonTorrent (Page 37, Figure 2) use that same definition that you mentioned.
So, the insert size term is a little bit misleading, as it in fact corresponds to the full length of the template including both mates (and not to the “inserted” length between the two mates, as its name would suggest).
But for 454 I couldn’t find any information about this…
Do you know if it’s the same way?
Thank you very much!
I tend to agree that ‘insert size’ is a bit misleading. The use of ‘insert size’ probably stems from the time when plasmids/fosmids and BACs were end-sequenced with Sanger. Then, insert size actually makes sense.
I am pretty sure for 454 the same definition is used, although I have no reference for it right now.
Hello!
In which step of the assembly do you specify the insert size? I assume you have to tell newbler what the insert sizes (3.5kb and 8.9kb) of the two datasets are. Is it possible to use other insert sizes?
kind regards,
Koen
You can’t tell newbler the insert size. It will determine it itself by mapping mates to contigs. It does mean one has to have sufficiently large contigs for this. See http://contig.wordpress.com/2010/02/09/how-newbler-works/ Any insert size is usefull, an important one has an insert size longer than the longest repeat.
Thanks for sharing data and code! I wonder why when calculating percentage of paired reads you divide number of unpaired non uniq reads by number of uniq reads? Why not to divide uniq by uniq or non_uniq by non_uniq?
I am dividing the number of unique, unpaired reads over the total number of unique reads to get the percentage of non-paired reads. The rest is then the % of paired reads…
But the first script calculates the sum of nonuniq reads. So 1557426 – is a number of nonuniq unpaired reads. Isn`t it?
No (and yes): it’s the number of all non-paired reads, each with a unique identifier. However, some of these reads may be duplicates of each other, – that is not taken into account. So, unique reads (wells on the chip), but not unique sequences.
Got it! My mistake
Dear lexnederbragt,
I followed the same protocol but i am not getting .fasta file in sffextract. Moreover, in case of fastq file the headings are not like .r and .f or \r and \f. I am getting \1 and \2. How to know which is forward and reverse?
Ex-
@IQVCN:00011:00045/1
@IQVCN:00011:00045/2
I have not used sffextract in a while. ‘/1’ and ‘/2’ stand for read 1 and read 2, respectively, with read 2 usually on the reverse strand. But you need to be careful and find out what orientation these paired reads are (–> <–, , or –> –>), and make sure you give your preferred assembly the reads in the right orientation.