As part of the bachelor-studies reform here at the University of Oslo, the Institute of Biosciences, where I work, is reorganising its bachelor curriculum. One exciting part is the implementation of the Computing in Science Education (CSE) project, into the different subjects. The goal of the CSE project is to make calculations/computing an integral part of the bachelor curriculum from the very first semester, interweaving the subject-specific material with mathematics and programming. CSE has been working successfully with the curriculum of the Physics, Mathematics, Informatics, Chemistry and Geology. Starting from the fall of 2017, Biology is next.
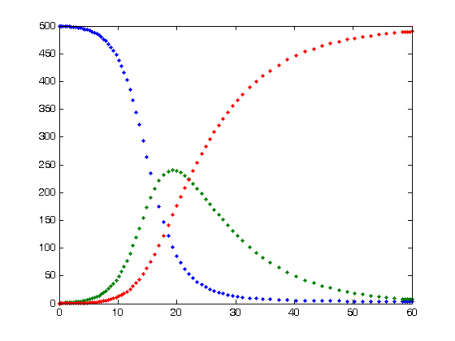
The SIR model for infectious diseases. number of Susceptible (blue=, Infected (green), and Recovered (red), over units of time. From Wikiemedia Commons
The Institute of Biosciences, with the CSE project, set itself an ambitious goal: in the first semester for Biosciences students, one of the three parallel 10-credit courses should be focussing on the CSE aspect. We named the course ‘Introduction to Computational Modelling for the Biosciences”. Four smart PhD students, with backgrounds in physics. informatics and biology, were tasked with writing a book for the course, in which they mix biology with programming in python and modeling of biological phenomena.
The CSE project and the institute have given me the opportunity (and honour) of developing this book, with the authors and the CSE project team, into a full first semester course for Biosciences students. This course (BIOS1100) is starting this fall (August 2017).
Wow. No small task. How to make sure 150 students (perhaps more) become, and then stay, motivated to learn programming in python and developing mathematical models, while what they really signed up for was studying biology! How to make sure the material appears relevant, and is seen as necessary for becoming a biologist? In preparing for teaching the course I have landed on a number of principles I want to adhere to, and ideas that I want to try, to ensure the course will be a success. Some of these are based on experiences obtained from previous courses developed within the CSE framework, as well as on my previous teaching experience. A significant part is based on my experience as a Software Carpentry instructor and instructor-trainer.
Ground the material in biology
Examples and exercises should be using data and questions that the students can relate to, given that they are studying biology/biosciences. As an example, another ‘CSE’ course uses a formula to calculate the time it takes for the centre of the yolk of an egg to reach a certain temperature. The students are asked to implement this formula in python, and calculate the time for the yolk to reach, say 70 C, when the egg is taken from a refrigerator into boiling water. I can still use the same formula, but would place the exercise in the setting of a chicken farmer, that would like to know how long a hen can be away from a fertilised egg before the temperature of the embryo becomes too low.
Build bridges with other courses
The two courses the students are taking in the same semester are Cell- and Molecular Biology, and Introduction to Chemistry. I have a very fruitful dialogue with one of the main responsible teachers for the Cell- and Molecular Biology course about ways to integrate what the students learn there, into the Modelling course. We are looking for opportunities to use small python programs to visualise or further analyse data they generate in the lab or are exposed to during the lectures. One of our plans that I am most looking forward to is what we put together for the subject of restriction digestion of DNA. Certain enzymes are able to recognise short DNA sequences and then make a break into the DNA at those sequences. Here is the plan:
- have students find the recognition sequence in DNA printed on a piece of paper and cut it with a pair of scissors (active learning, pen-and-paper exercise)
- have students make a small script that cuts a DNA sequencing in silico given a recognition site
- have students cut three real DNA (plasmid) sequences using their program and predict the sizes of the resulting fragments (ideally, they would simulate the visualisation of the results of electrophoretically separating the fragments on a so-called agarose gel)
- in the laboratory, have students cut the DNA of the three actual plasmids with the restriction enzyme, without telling them which plasmid is which
- have students determine which restriction digest corresponds to which input plasmid DNA sequence
My hope is that by exposing students to the subject in multiple ways, on multiple occasions, learning about restriction digestion becomes more effective (and perhaps also more fun). I also am looking into the labwork the students will do in the parallel courses to see whether they generate any datasets that can be further analysed using python or modelling. I suspect that working with data students generate themselves (i.e., have ownership over) enhances motivation and increases learning outcome.
Use methods and create an environment that enhance learning
One cannot learn programming from slide-based lectures. Much of the learning will have to be done ‘by doing’, in group work. I will use the Jupyter Notebook, the “killer app” in education according to professor Lorena Barba, in a flipped classroom approach where students study notebooks beforehand (each chapter of the course book can be turned into a notebook), formative assessment is used to gauge understanding, and students work with exercises during ‘class’. We have the fortunate situation that we could design a new classroom to be used for this and others courses. Inspired by a teaching room at the University of Minnesota and this publication we will fill it with hexagonal tables that each have their own large screen for students to share. We will be using a ‘bring-your-own-device’ approach (students working on their own laptops) rather than through Data Labs with stationary, university PCs. This ensures learning can continue on the students own equipment, wherever they are, whenever they want to.
Build a community of learning assistants
I need to recruit quite number of learning assistants, PhD students and others that work with the students during group work. The aim is one assistant per two hexagonal tables (12 students). How they interact with the students will influence the learning and student experience significantly. I aim to have a couple of small workshops with them before the course starts (using some of the Software and Data Carpentry Instructor Training material for inspiration). During the course we will need to meet weekly to discuss what went well and where we can improve. We will also need to document all this for the next editions of the course.
Trial course
These weeks, we are trying out some of the material on a small group of current first year bachelor students. This turns out to be very useful, it makes us think about many aspects that otherwise first would have become apparent during the real course. For example, I had first thought to do quite a bit of learning through live-coding, Software Carpentry style. However, I have come to realise that live-coding does not scale: classes will be 50-60 students rather than the typical 20-30 for a Software or Data Carpentry workshop. The chances for delays due to a student getting into a technical problem are much higher, and the number of people affected by the delay much larger. This will cause problems to keep the group’s attention and may become demotivating.
Challenges
A major challenge will be the motivational aspect, and we should get feedback from the students on this and other aspects frequently. I have lot’s of ideas on what to try to improve the students’ learning and experience but will need to limit myself this year to getting through the course. Finally, the overall aim is not to have this work stop with this one single course. The goal is that all other Biosciences courses incorporate modelling with python, and also R, in group and project-based work. This requires the collaboration of all teachers in the department. At a departmental teaching seminar last year everyone was very enthusiastic. Soon is the time to turn that enthusiasm into concrete projects.
Some more reading
In English:
- The CSE crew was recently awarded a Center for Excellence in Education, see the website of the Centre for Computing in Science Education (CCSE)
In Norwegian:

That sounds challenging, but misguided. Biology is not a model-driven field, but a data-driven one, and biology students would be better off learning to program to handle data than to run simulations. The BME 160 course at UCSC is probably a better course for biology students, though I can’t find a current syllabus or set of assignments on the web to point to for explaining the course.
Thanks for your comment (I have been following your blog for a while). I partly agree, and partly think the post could have described something better. First-year biology students used to have a mathematics course (10 credits) in their first semester, and this course is partly meant as a replacement. I agree in that biology is currently much data-driven, and this and other courses in the curriculum should indeed train students data analysis and handling. But I do also think that biology needs more, better and more complex models if we ever want to be able to make sense out of all that ‘omics-‘data we are now able to generate.
Biology may need better models, but I doubt that they will come from studying the traditional differential equations and predator-prey models. I think that Bayesian statistics and factor graphs may be more productive, if only because they emphasize the importance of stochastic, rather than deterministic, modeling.
I am a recent graduate from the University of Minnesota’s College for Biological Sciences, and one of my favorite classes (and one where I learned the most) was a flipped classroom course in genetics in the active learning classrooms. Its cool to learn that these concepts are getting implemented more. This sounds like a great project and I wish you luck!
Thanks!