In 1986, in a letter to the journal Nature, James Bruce Walsh and Jon Marks lamented that the upcoming human genome sequencing project “violates one of the most fundamental principles of modern biology: that species consist of variable populations of organisms”. They further wrote: “As molecular biologists generally ignore any variability within a population, the individual whose haploid [sic] genome will be chosen will provide the genetic benchmark against which deviants are determined”. They conclude that ” ‘the’ genome of ‘the’ human will be sequenced gel by acrylamide gel”.
We have come a long way when it comes to taking population variation into account in molecular/genetic/genomic studies. But these sentiments, expressed already in 1986, echo some of the trends in the human genetics field: the move away from a single, linear representation of ‘the’ human genome. In this post I will provide some background, explain the reasons for moving towards graph-based representations, and indicate some challenges associated with this development.
Reference genome as a line
Traditionally, reference genomes are represented as one or multiple sequences, interrupted with strings of ‘N’s to indicate gaps if the reference is incomplete. Graphically, the sequences are represented as straight lines. Annotations are represented as genomic tracks. A paper by Gundersen et al identified 15 different track types.
Limitations of the linear representation
Diploid organisms receive two sets of chromosomes, one from each
parent. The sequences of these chromosomes are similar (technically they are ‘homologous’), but have many differences (so-called ‘heterozygosities’). These variations can be small, such as Single Nucleotide Variants (SNVs, also known as Single Nucleotide Polymorphisms, SNPs), small insertions and deletions (collectively known as Indels), copy number variations; they can also span larger distances, such as chromosomal rearrangements (deletions, duplications, inversions, and translocations). To represent this diploid genome as a single linear sequence necessarily removes some of this variation, or requires an additional way of reporting the variations of the individual chromosomes relative to the linear representation. A complicating factor is that every new studied instance of a genome may contain sequences not represented in the reference (see for example the paper by Liu et al and references therein). When sequencing a new individual, and mapping the reads to the linear representation of the reference, the parts of the sequenced sample not present in the reference may thus be missed.
Solutions that are not graph-based
The most recent release of the human genome (build 38, GRCh38) contains so-called alternate loci, defined as “multiple representations for regions that are too complex to be represented by a single path”. These are regions, at most 1Mbp (million base pair), that are found in some genomes, but are not part of the default reference representation (see the official website for the
human genome). Read mapping software is being developed that takes these alternate loci into consideration, allowing for more reads to be mapped to the right position, thus reducing false mappings, and false positive variant calls. The popular read mapper bwa has been made ‘alt-aware’ to cope with these alternate loci. There also is an unpublished read mapper called SRPRISM mentioned in a preprint paper on a haploid human genome assembly.
Graph-based representation as a solution
Using graphs to represent DNA sequences has a long history. Most, if not all, assembly programs use graph representations and graph algorithms to assemble reads into a genome representation. Both String Graphs (e.g., Myers et al 2005), and De Bruijn graphs (see e.g. [Compeau et al 2011(http://www.nature.com/nbt/journal/v29/n11/full/nbt.2023.html)) are used in this context. Representing a diploid genome as a graph allows for alternative paths (‘bubbles’) for regions where the chromosomes differ.
But a graph could also be used to represent the genomes of multiple individuals, capturing all variation between them, and potentially containing all sequences found in the represented organisms. The graph could have ‘colored’ paths, where parts of the graph are annotated with the sample(s) they represent. A ‘colored de Bruijn graph’ approach has been used for the global ‘superassembly’ program cortex, or for the local ‘variant-calling through assembly’ tool platypus. A different approach is taken by the (unpublished, but openly developed) program vg (variant graph). This program starts from a reference genome, and a vcf file of variants between the reference and one or more samples, and then builds a graph to which reads can be mapped.
Potential gains
Having a graph to represent all that is known about a genome would allow for a higher percentage of reads of a new instance of the genome to be mapped correctly. It is probably easier to map reads corresponding to variants if those variants are present in the reference graph (e.g. indels, structural variations). The example below shows two different alignments of the same variant read to a reference:
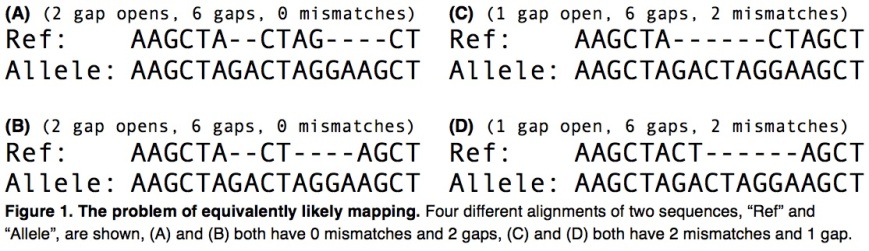
The problem of equivalent mappings. Image source: Paten, Benedict, Adam Novak, and David Haussler (2014) arXiv:1404.5010
Source: Paten, Benedict, Adam Novak, and David Haussler (2014) arXiv:1404.5010
Also, graph-based representations enable new ways to integrate multiple different ‘-omics’ datatypes. For example, coupling genetic variation with epigenetic information (bases methylated in some organisms may be absent from the genomes of other individuals).
What to represent in the graph
There are several different possibilities to represent genomes with a graph:
- the genome of a single diploid (or polypoid) individual, for example after assembly. For this, a ‘diploid aware assembler’ would be needed. There are developments towards this goal, see for example the experimental PacBio diploid assembler FALCON
- the genomes of multiple individuals of the same species
- based on reads: superassembly
- based on multiple (complete) reference genomes, e.g. for multiple strains of bacteria
- the genomes of multiple species
- building a graph to represent multiple reference genomes for comparative genomics
Challenges
A few of the challenges of using a graph to represent a genome are
briefly discussed here.
- a linear representation allows an unambiguous coordinate system: sequence X at position Y has this base, or this feature. Defining distances between features is then very straightforward (e.g. the transcription binding factor recognition site is located Z bases upstream of the gene). A graphical representation makes unambiguous coordinates challenging. For regions with alternative paths, new ways to define coordinates (absolute, relative) are needed. The concept of proximity, i.e. ‘distance between features’, may have to be defined in other (statistical?) terms
- similarly, annotations (genomic tracks) currently simply refer to the linear genome representation in a one-to-one fashion: coordinates on the track correspond to coordinates on the reference genome. With a graph, in certain parts of the genome the annotation will be for not just one path, but several (variant) paths. The annotation would need to refer to the entire subgraph that represents the feature. For example, to annotation gene X, the collective set of paths through the graph at the ‘locus’ of gene X must be annotated as such
- in many cases, it is useful to randomize annotations. One may for instance randomize the location of genes, and compare some characteristics of the real gene locations with the randomized ones. With a linear genome, this is essentially just to draw a random number (position on the line) for each gene. For a graph, it is not apparent how one should take the alternative paths into account when randomizing for example positions
- a graph representing the genome of a species will grow (be updated) as more data on that species becomes available. Strategies to ‘lift over’ findings from earlier versions need to be developed
- file formats to represent the graph are needed, but they would preferably be based on/compatible with existing community standards, or those standards should be easily derived from them
- mapping to the graph instead of a linear reference genome requires specialized software
- the current most popular visualisation of genomes and their annotation is through genome browsers. These work very well with linearly represented sequences and annotation tracks. The challenge is to visualise a graphical representation in an intuitive way
- closer to home, the hyperbrowser statistical genomics framework was developed at the University of Oslo around the concept of the genomic track (the hyperbrowser slogan is “If you have a genomic track, this is the place to analyze it!”). A graphical representation of a genome adds a layer of complexity, at the same time opening for new way of doing statistics on (groups of) genomes
- combining reference genomes of multiple species into a single graph requires handling of (imperfect or non-equivalent) annotations as well as structural variations between the chromosomes
Prior work
The concept of representing (reference) genomes as graphs is attracting more and more attention. There are a few relevant published papers, see the bibliography at the end. Heng Li, author of, among other programs, BWA and Samtools has written an excellent summary blog post in which he succinctly summarises the material, including prior work from computer science and computational biology.
Two file formats to represent the graph been developed: fastg and GFA. Fastg has limited uptake, only two assembly programs (ALLPATHS_LG and SPAdes) will output in that format. GFA parsing is currently only experimentally in the ABYSS assembler, and also vg mentioned above is able to output it.
Why are we interested in Graph-based representations of reference genomes
* we are involved in projects sequencing many individuals of the same species. For example, in the AquaGenome project we will obtain short-read sequencing data for 1000 individuals of Atlantic cod. Making sense of this data will be helped hugely if we can use graph-based methods instead of a single linear reference genome
* other projects we contribute to will generate assemblies for different species with the aim of doing comparative genomics. We are interested in exploring graph-based methods also in this area
* the Genomic hyperbrowser group has many tools for comparing genomic tracks, and is interested in developing their analysis platform in the light of graph-based reference genomes
Conclusion
‘The’ genome of ‘the’ human has been sequenced, gel by acrylamide gel, and new human genomes are being resequenced, flowcell by flowcell. Many organisms have similar reference genomes available now, and deviants are being determined against these genetic benchmarks. But my hope is that in the not too distance future, we’ll have ways to truly represent the genomes of the ‘variable populations of organisms’ that Walsh and Marks rightly consider so fundamental for our understanding of biology.
Recent papers of relevance
The UCSC genome browser group (Haussler D, Paten B) uploaded a few
highly relevant papers to arXiv in 2014 and 2015:
- Paten, Benedict, Adam Novak, and David Haussler. “Mapping to a reference genome structure.” arXiv:1404.5010 (2014).
- Novak, Adam et al. “Canonical, Stable, General Mapping using Context Schemes.” arXiv:1501.04128 (2015)
Other important recent papers:
- Dilthey, Alexander et al. “Improved genome inference in the MHC
using a population reference graph.” bioRxiv (2014): 006973. - Nguyen, Ngan et al. “Building a pangenome reference for a population.” Research in Computational Molecular Biology (2014): 207-221.
- Church, Deanna M et al. “Extending reference assembly models.” Genome biology 16.1 (2015): 13.
Further reading
See the aforementioned summary blog post by Heng Li with additional references from computer science and computational biology.

Hi Lex, just to add to the many awesome links above, this also seems relevant http://biorxiv.org/content/early/2016/03/12/043430
Thanks!