Pacific Biosciences published a paper earlier this year on an approach to sequence and assemble a bacterial genome leading to a near-finished, or finished genome. The approach, dubbed Hierarchical Genome Assembly Process (HGAP), is based on only PacBio reads without the need for short-reads. This is how it works:
- generate a high-coverage dataset of the longest reads possible, aim for 60-100x in raw reads
- pre-assembly: use the reads from the shorter part of the raw read length distribution, to error-correct the longest reads, set the cutoff in such a way so that the longest reads make up about 30x coverage
- use the long, error-corrected reads in a suitable assembler, e.g. Celera, to produce contigs
- map the raw PacBio reads back to the contigs to polish the final sequence (rather, recall the consensus using the raw reads as evidence) with the Quiver tool
The approach is very well explained on this website. As an aside, the same principle can now be used with the PacBioToCA pipeline.
In principle, this approach could result in a finished genome, i.e. a gapless contig per chromosomal element (chromosomes and plasmids). A more theoretical study confirms this:
“Our results indicate that the majority of known bacterial and archaeal genomes can be assembled without gaps, at finished-grade quality, using a single PacBio RS sequencing library.” (Koren et al, arXiv:1304.3752)
As always, the proof is in the pudding. There have been reports, and even a publication here and there, that the HGAp approach actually works. In this blog post I would like to add our experiences, which in short are that HGAP can indeed result in (close-to) finished genomes.
At the Norwegian Sequencing Centre,with which I am affiliated, we recently received several bacterial genome DNA samples for PacBio sequencing. Given our very positive first experiences with size selecting PacBio libraries using the BluePippin, see my previous post, we decided to use this instrument also for these samples. Four of the samples yielded very nice libraries, which were sequenced, two SMRTcells each, on our (recently upgraded) PacBio RSII instrument.
Raw reads
We have never seen such long reads:
| PB_0027 | PB_0028 | PB_0029 | PB_0031 | |
| Count | 80512 | 58524 | 45514 | 84169 |
| Sum | 595 Mbp | 462 Mbp | 351 Mbp | 669 Mbp |
| Av. (bp) | 7393 | 7893 | 7714 | 7951 |
| N50 (bp) | 10662 | 11205 | 11109 | 11162 |
| Largest (bp) | 24397 | 25552 | 23992 | 25678 |
Note that the average read length is much longer than the specifications of the RSII, which is about 4.6 kbp.
These reads were then used in HGAP. We have smrtpipe, the analysis suite of Pacific Biosciences, installed, so I could simply make a file with the names of the input files, a default HGAP settings xml file, and run the whole thing on one of our big servers. The assemblies took about two days when given 32 CPUs and a lot of memory – I haven’t logged how much RAM they actually used.
Pre-assembly
Here are the results of pre-assembly, the correction of the largest 30x in raw reads with the rest of the reads:
| PB_0027 | PB_0028 | PB_0029 | PB_0031 | |
| Cutoff (bp)* | 12106 | 12077 | 10371 | 12780 |
| Count | 9186 | 8636 | 10059 | 9252 |
| Sum | 107 Mbp | 100 Mbp | 106 Mbp | 110 Mbp |
| Av. (bp) | 11594 | 11562 | 10540 | 11876 |
| N50 (bp) | 12519 | 12770 | 11513 | 13120 |
| Largest | 20043 | 19090 | 19030 | 18681 |
*Cutoff: minimum length of seeds for error-correction.
After pre-assembly, there was more than a 100 Mbp in error-corrected, potentially high-quality reads with an N50 higher than one sometimes see for contigs of a short-read bacterial genome assembly!
Assembly
These 8 – 10 thousand reads were assembled by Celera, with Quiver polishing, into:
| PB_0027 | PB_0028 | PB_0029 | PB_0031 | |
| Contigs | 3.4 Mbp | 3.2 Mbp | 4.3 Mbp | 1.8 Mbp |
| 45 kbp | 76 kbp | 80kbp | 1.3 Mbp | |
| 64 Kbp | 1.1 Mbp | |||
| 45 Kbp | 0.95 Mbp | |||
| 17 kbp | 95 kbp |
Wow, mostly one laaarge contig (and I checked, these are without ‘N’ bases) and a few shorter ones. The exception was the last strain which assembled into a few large pieces, that together, according to what I understand, are too large. A further step for this assembly is trying the Minimus2 tool, to see whether there is enough overlap between the contigs to further reduce their number – a step generally recommended for HGAp assemblies. I haven’t tried this yet for this assembly.
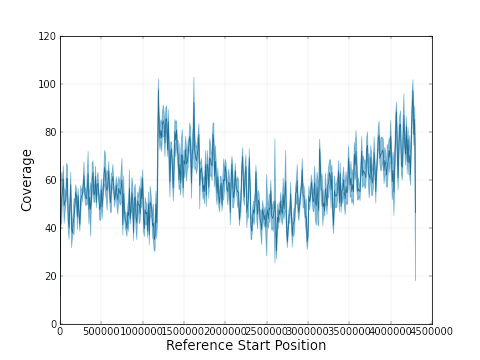
So, it looks like ‘it just works’. Well, there was at least one case where a misassembly is suspected. Looking at the coverage plot (of the remapped raw pacbio reads) for the 4,3 Mbp contig of PB0029, we saw this:
The sudden jump in coverage after 1.2 Mbp points to a fusion of the sequences of two chromosomes – and in fact this is quite likely the case given what is know about these strains. For the others, it reamins to be seen whether the smaller pieces are in fact plasmids, or should be part of the major chromosome.
A few remarks before I conclude:
- these four samples are clearly success stories
- all had modest GC percentages, around 35 – 50%
- we also have had a sample that didn’t fragment very well and only yielded a 2 kbp insert library (giving CCS reads after sequencing)
- another strain didn’t behave as well either, resulting in reads averaging 3.5 kbp – assembly for this one has not been started yet
- there is no reference genome for these samples, so assembly accuracy, and per-base quality, could not be assessed fully
Conclusion
It looks like that for well-behaved samples, the approach of combining PacBio library creation, BluePippin size selection (optional, but highly recommended) and sequencing of two SMRTcells, works very well to give finished, or near finished bacterial genome assemblies. I want to emphasise the following, though: even though the assembly looks great, it is afterwards up to the biologist/researcher to make sure the contigs actually make sense given:
- the remapping of the reads
- what is known about the species (e.g. expected number of chromosomes)
- what is known about the sample (e.g., presence of plasmids)
- other, independent evidence, e.g. illumina reads, optical mapping results, etc
The title of this post aks: “De novo bacterial genome assembly: a solved problem?”. I dare to say we’re pretty close…
A bioinformaticians side-note
The bottleneck of the HGAp process was the two consensus calling steps: when the consensus of the longest reads are being called (based on the mapped shortest ones), and especially for the Celera contig consensus calling. The latter takes one contig at a time, and since these now are becoming millions of basepairs long, this can take up many hours, perhaps even half the total assembly time. By the way, overlapping the error-corrected reads was done in minutes… So, if someone is interested in developing a parallelised consensus caller, than can work with parts of a long contigs, and stitch the consensi back together when done, we bioinformaticians doing HGAP would be very grateful…
Acknowledgments
This post would not have been possible without the excellent skills of the NSC lab team, and I thank the owners of the Bacterial samples for which this post describes results for permission to use the metrics for this post. I apologize in advance for not being able to share the (raw and assembled) data presented here…

Very nice results. I admit – I’m jealous. Now I just need to find the $700K for the instrument! 🙂
Jonathon: don’t buy, rent! Many core labs (including Lex’s) run samples from all comers at reasonable rates
Yes, there are definitely several options for researchers to explore PacBio sequencing. Here is a directory page of sequencing providers accepting external projects:
http://www.pacificbiosciences.com/support/sequencing_provider/
I finally had the time to read your post. Interesting results.
So I am a bit concerned about the fragmentation of the DNA that is needed to create the libraries because of my own project. You mention that with some strains the fragmentation did not go well and gave only relatively short reads. Would you then still consider the same approach using 2 SMRT cells, or would you try something else to optimize your assemblies?
If the fragmentation doesn’t yield the very long fragments needed for a successful HGAP-based assembly, the best one can do – besides starting from scratch – is still giving it a try and see how far the assembly goes. It may result in many more contigs, which could be scaffolded with, for example, mate pairs, or comparing to a closely related, finished genome.